About Ral GTPase
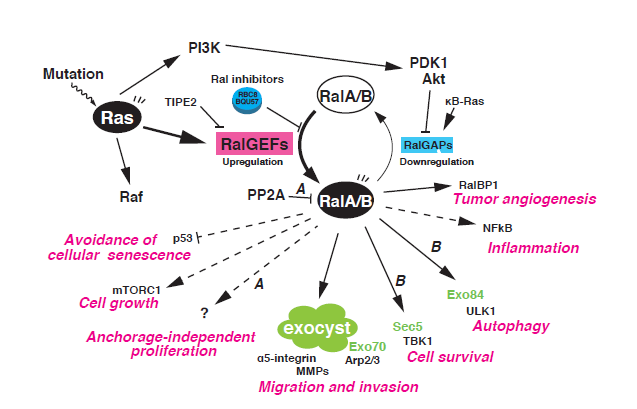 "The Ral GTPases, RalA and RalB, are members of the Ras superfamily of small GTPases. Research on Ral GTPases and their functions over the past 25 years has revealed the essential involvement of these GTPases in unique and diverse cellular processes including exocyst-mediated exocytosis and related cellular activities. Moreover, it is increasingly appreciated that the aberrant activation of Ral GTPases is one of the major causes of human tumorigenesis induced by oncogenic Ras. Recent evidence suggests that Ral signaling pathways may be potential therapeutic targets for the treatment of human cancers."
(Shirakawa and Horiuchi, J Biochemistry, 2015)
"The Ral GTPases, RalA and RalB, are members of the Ras superfamily of small GTPases. Research on Ral GTPases and their functions over the past 25 years has revealed the essential involvement of these GTPases in unique and diverse cellular processes including exocyst-mediated exocytosis and related cellular activities. Moreover, it is increasingly appreciated that the aberrant activation of Ral GTPases is one of the major causes of human tumorigenesis induced by oncogenic Ras. Recent evidence suggests that Ral signaling pathways may be potential therapeutic targets for the treatment of human cancers."
(Shirakawa and Horiuchi, J Biochemistry, 2015)
|
Identification of RalGAPs
  "The small GTPases RalA and RalB are multifunctional pro-
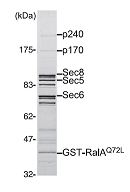
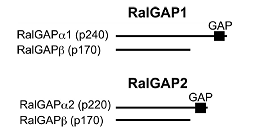
teins regulating a variety of cellular processes. Like other GTPases, the activity of Ral is regulated by the opposing effects of guanine nucleotide exchange factors (GEFs) and GTPase-ac- tivating proteins (GAPs). Although several RalGEFs have been identified and characterized, the molecular identity of RalGAP remains unknown. We reported the first molecular identification of RalGAPs, which we have named RalGAP1 and Ral- GAP2. They are large heterodimeric complexes, each consisting of a catalytic α1 or α2 subunit and a common β subunit. These RalGAP complexes share structural and catalytic similarities with the tuberous sclerosis tumor suppressor complex, which acts as a GAP for Rheb. In vitro GTPase assays revealed that recombinant RalGAP1 accelerates the GTP hydrolysis rate of RalA by 280,000-fold. Heterodimerization was required for this GAP activity. In PC12 cells, knockdown of the βsubunit led to sustained Ral activation upon epidermal growth factor stimulation, indicating that the RalGAPs identified here are critical for efficient termination of Ral activation induced by extracellular stimuli. Our identification of RalGAPs will enable further understanding ofRal signaling in manybiological and pathological processes." (Shirakawa et al, J Bio Chem, 2009)
"The small GTPases RalA and RalB are multifunctional pro-
teins regulating a variety of cellular processes. Like other GTPases, the activity of Ral is regulated by the opposing effects of guanine nucleotide exchange factors (GEFs) and GTPase-ac- tivating proteins (GAPs). Although several RalGEFs have been identified and characterized, the molecular identity of RalGAP remains unknown. We reported the first molecular identification of RalGAPs, which we have named RalGAP1 and Ral- GAP2. They are large heterodimeric complexes, each consisting of a catalytic α1 or α2 subunit and a common β subunit. These RalGAP complexes share structural and catalytic similarities with the tuberous sclerosis tumor suppressor complex, which acts as a GAP for Rheb. In vitro GTPase assays revealed that recombinant RalGAP1 accelerates the GTP hydrolysis rate of RalA by 280,000-fold. Heterodimerization was required for this GAP activity. In PC12 cells, knockdown of the βsubunit led to sustained Ral activation upon epidermal growth factor stimulation, indicating that the RalGAPs identified here are critical for efficient termination of Ral activation induced by extracellular stimuli. Our identification of RalGAPs will enable further understanding ofRal signaling in manybiological and pathological processes." (Shirakawa et al, J Bio Chem, 2009)
|
The function of Ral in cancer
Bladder cancer
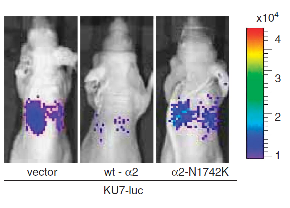 "The small GTPase Ral is known to be highly activated in several human cancers, such as bladder, colon and pancreas cancers. It is reported that activated Ral is involved in cell proliferation, migration and metastasis of bladder cancer. This protein is activated by Ral guanine nucleotide exchange factors (RalGEFs) and inactivated by Ral GTPase-activating proteins (RalGAPs), the latter of which consist of heterodimers containing a catalytic a1or a2 subunit and a common b subunit. In Ras-driven cancers, such as pancreas and colon cancers, constitutively active Ras mutant activates Ral through interaction with RalGEFs, which contain the Ras association domain. However, little is known with regard to the mechanism that governs aberrant activation of Ral in bladder cancer, in which Ras mutations are relatively infrequent. Here, we show that Ral was highly activated in invasive bladder cancer cells due to reduced expression of RalGAPa2, the dominant catalytic subunit in bladder, rather than increased expression of RalGEFs. Exogenous expression of wild-type RalGAPa2 in KU7 bladder cancer cells with invasive phenotype, but not mutant RalGAPa2-N1742K lacking RalGAP activity, resulted in attenuated cell migration in vitro and lung metastasis in vivo. Furthermore, genetic ablation of Ralgapa2 promoted tumor invasion in a chemically-induced murine bladder cancer model. Importantly, immunohistochemical analysis of human bladder cancer specimens revealed that lower expression of RalGAPa2 was associated with advanced clinical stage and poor survival of patients. Collectively, these results are highly indicative that attenuated expression of RalGAPa2 leads to disease progression of bladder cancer through enhancement of Ral activity." (Saito et al, Oncogene, 2013)
"The small GTPase Ral is known to be highly activated in several human cancers, such as bladder, colon and pancreas cancers. It is reported that activated Ral is involved in cell proliferation, migration and metastasis of bladder cancer. This protein is activated by Ral guanine nucleotide exchange factors (RalGEFs) and inactivated by Ral GTPase-activating proteins (RalGAPs), the latter of which consist of heterodimers containing a catalytic a1or a2 subunit and a common b subunit. In Ras-driven cancers, such as pancreas and colon cancers, constitutively active Ras mutant activates Ral through interaction with RalGEFs, which contain the Ras association domain. However, little is known with regard to the mechanism that governs aberrant activation of Ral in bladder cancer, in which Ras mutations are relatively infrequent. Here, we show that Ral was highly activated in invasive bladder cancer cells due to reduced expression of RalGAPa2, the dominant catalytic subunit in bladder, rather than increased expression of RalGEFs. Exogenous expression of wild-type RalGAPa2 in KU7 bladder cancer cells with invasive phenotype, but not mutant RalGAPa2-N1742K lacking RalGAP activity, resulted in attenuated cell migration in vitro and lung metastasis in vivo. Furthermore, genetic ablation of Ralgapa2 promoted tumor invasion in a chemically-induced murine bladder cancer model. Importantly, immunohistochemical analysis of human bladder cancer specimens revealed that lower expression of RalGAPa2 was associated with advanced clinical stage and poor survival of patients. Collectively, these results are highly indicative that attenuated expression of RalGAPa2 leads to disease progression of bladder cancer through enhancement of Ral activity." (Saito et al, Oncogene, 2013)
Oral squamous cell carcinoma
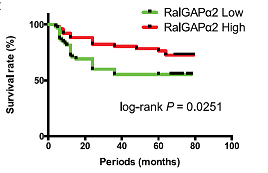 "Abstract
Ral small GTPases, consisting of RalA and RalB, are members of the Ras family. Their activity is upregulated by RalGEFs. Since several RalGEFs are downstream effectors of Ras, Ral is activated by the oncogenic mutant Ras. Ral is negatively regulated by RalGAP complexes that consist of a catalytic α1 or α2 subunit and its common partner β subunit and similarly regulate the activity of RalA as well as RalB in vitro. Ral plays an important role in the formation and progression of pancreatic and lung cancers. However, the involvement of Ral in oral squamous cell carcinoma (OSCC) is unclear. In this study, we investigated OSCC by focusing on Ral. OSCC cell lines with high Ral activation exhibited higher motility. We showed that knockdown of RalGAPβ increased the activation level of RalA and promoted the migration and invasion of HSC-2 OSCC cells in vitro. In contrast, overexpression of wild-type RalGAPα2 in TSU OSCC cells attenuated the activation level of RalA and inhibited cell migration and invasion. Real-time quantitative polymerase chain reaction analysis of samples from patients with OSCC showed that RalGAPα2 was downregulated in oral cancer tissues as compared with normal epithelia. Among patients with OSCC, those with a lower expression of RalGAPα2 showed a worse overall survival rate. A comparison of DNA methylation and histone modifications of the RalGAPα2 gene in OSCC cell lines suggested that crosstalk among DNA methylation, histone H4Ac, and H3K27me2 was involved in the downregulation of RalGAPα2. Thus, activation of Ral GTPase by downregulation of RalGAP expression via a potential epigenetic mechanism may enhance OSCC progression.
Keywords:." (Gao et al, J Dentarl Res, 2019)
"Abstract
Ral small GTPases, consisting of RalA and RalB, are members of the Ras family. Their activity is upregulated by RalGEFs. Since several RalGEFs are downstream effectors of Ras, Ral is activated by the oncogenic mutant Ras. Ral is negatively regulated by RalGAP complexes that consist of a catalytic α1 or α2 subunit and its common partner β subunit and similarly regulate the activity of RalA as well as RalB in vitro. Ral plays an important role in the formation and progression of pancreatic and lung cancers. However, the involvement of Ral in oral squamous cell carcinoma (OSCC) is unclear. In this study, we investigated OSCC by focusing on Ral. OSCC cell lines with high Ral activation exhibited higher motility. We showed that knockdown of RalGAPβ increased the activation level of RalA and promoted the migration and invasion of HSC-2 OSCC cells in vitro. In contrast, overexpression of wild-type RalGAPα2 in TSU OSCC cells attenuated the activation level of RalA and inhibited cell migration and invasion. Real-time quantitative polymerase chain reaction analysis of samples from patients with OSCC showed that RalGAPα2 was downregulated in oral cancer tissues as compared with normal epithelia. Among patients with OSCC, those with a lower expression of RalGAPα2 showed a worse overall survival rate. A comparison of DNA methylation and histone modifications of the RalGAPα2 gene in OSCC cell lines suggested that crosstalk among DNA methylation, histone H4Ac, and H3K27me2 was involved in the downregulation of RalGAPα2. Thus, activation of Ral GTPase by downregulation of RalGAP expression via a potential epigenetic mechanism may enhance OSCC progression.
Keywords:." (Gao et al, J Dentarl Res, 2019)
|

 "The Ral GTPases, RalA and RalB, are members of the Ras superfamily of small GTPases. Research on Ral GTPases and their functions over the past 25 years has revealed the essential involvement of these GTPases in unique and diverse cellular processes including exocyst-mediated exocytosis and related cellular activities. Moreover, it is increasingly appreciated that the aberrant activation of Ral GTPases is one of the major causes of human tumorigenesis induced by oncogenic Ras. Recent evidence suggests that Ral signaling pathways may be potential therapeutic targets for the treatment of human cancers."
(Shirakawa and Horiuchi, J Biochemistry, 2015)
"The Ral GTPases, RalA and RalB, are members of the Ras superfamily of small GTPases. Research on Ral GTPases and their functions over the past 25 years has revealed the essential involvement of these GTPases in unique and diverse cellular processes including exocyst-mediated exocytosis and related cellular activities. Moreover, it is increasingly appreciated that the aberrant activation of Ral GTPases is one of the major causes of human tumorigenesis induced by oncogenic Ras. Recent evidence suggests that Ral signaling pathways may be potential therapeutic targets for the treatment of human cancers."
(Shirakawa and Horiuchi, J Biochemistry, 2015)

 "The small GTPases RalA and RalB are multifunctional pro-
teins regulating a variety of cellular processes. Like other GTPases, the activity of Ral is regulated by the opposing effects of guanine nucleotide exchange factors (GEFs) and GTPase-ac- tivating proteins (GAPs). Although several RalGEFs have been identified and characterized, the molecular identity of RalGAP remains unknown. We reported the first molecular identification of RalGAPs, which we have named RalGAP1 and Ral- GAP2. They are large heterodimeric complexes, each consisting of a catalytic α1 or α2 subunit and a common β subunit. These RalGAP complexes share structural and catalytic similarities with the tuberous sclerosis tumor suppressor complex, which acts as a GAP for Rheb. In vitro GTPase assays revealed that recombinant RalGAP1 accelerates the GTP hydrolysis rate of RalA by 280,000-fold. Heterodimerization was required for this GAP activity. In PC12 cells, knockdown of the βsubunit led to sustained Ral activation upon epidermal growth factor stimulation, indicating that the RalGAPs identified here are critical for efficient termination of Ral activation induced by extracellular stimuli. Our identification of RalGAPs will enable further understanding ofRal signaling in manybiological and pathological processes." (Shirakawa et al, J Bio Chem, 2009)
"The small GTPases RalA and RalB are multifunctional pro-
teins regulating a variety of cellular processes. Like other GTPases, the activity of Ral is regulated by the opposing effects of guanine nucleotide exchange factors (GEFs) and GTPase-ac- tivating proteins (GAPs). Although several RalGEFs have been identified and characterized, the molecular identity of RalGAP remains unknown. We reported the first molecular identification of RalGAPs, which we have named RalGAP1 and Ral- GAP2. They are large heterodimeric complexes, each consisting of a catalytic α1 or α2 subunit and a common β subunit. These RalGAP complexes share structural and catalytic similarities with the tuberous sclerosis tumor suppressor complex, which acts as a GAP for Rheb. In vitro GTPase assays revealed that recombinant RalGAP1 accelerates the GTP hydrolysis rate of RalA by 280,000-fold. Heterodimerization was required for this GAP activity. In PC12 cells, knockdown of the βsubunit led to sustained Ral activation upon epidermal growth factor stimulation, indicating that the RalGAPs identified here are critical for efficient termination of Ral activation induced by extracellular stimuli. Our identification of RalGAPs will enable further understanding ofRal signaling in manybiological and pathological processes." (Shirakawa et al, J Bio Chem, 2009)
 "The small GTPase Ral is known to be highly activated in several human cancers, such as bladder, colon and pancreas cancers. It is reported that activated Ral is involved in cell proliferation, migration and metastasis of bladder cancer. This protein is activated by Ral guanine nucleotide exchange factors (RalGEFs) and inactivated by Ral GTPase-activating proteins (RalGAPs), the latter of which consist of heterodimers containing a catalytic a1or a2 subunit and a common b subunit. In Ras-driven cancers, such as pancreas and colon cancers, constitutively active Ras mutant activates Ral through interaction with RalGEFs, which contain the Ras association domain. However, little is known with regard to the mechanism that governs aberrant activation of Ral in bladder cancer, in which Ras mutations are relatively infrequent. Here, we show that Ral was highly activated in invasive bladder cancer cells due to reduced expression of RalGAPa2, the dominant catalytic subunit in bladder, rather than increased expression of RalGEFs. Exogenous expression of wild-type RalGAPa2 in KU7 bladder cancer cells with invasive phenotype, but not mutant RalGAPa2-N1742K lacking RalGAP activity, resulted in attenuated cell migration in vitro and lung metastasis in vivo. Furthermore, genetic ablation of Ralgapa2 promoted tumor invasion in a chemically-induced murine bladder cancer model. Importantly, immunohistochemical analysis of human bladder cancer specimens revealed that lower expression of RalGAPa2 was associated with advanced clinical stage and poor survival of patients. Collectively, these results are highly indicative that attenuated expression of RalGAPa2 leads to disease progression of bladder cancer through enhancement of Ral activity." (Saito et al, Oncogene, 2013)
"The small GTPase Ral is known to be highly activated in several human cancers, such as bladder, colon and pancreas cancers. It is reported that activated Ral is involved in cell proliferation, migration and metastasis of bladder cancer. This protein is activated by Ral guanine nucleotide exchange factors (RalGEFs) and inactivated by Ral GTPase-activating proteins (RalGAPs), the latter of which consist of heterodimers containing a catalytic a1or a2 subunit and a common b subunit. In Ras-driven cancers, such as pancreas and colon cancers, constitutively active Ras mutant activates Ral through interaction with RalGEFs, which contain the Ras association domain. However, little is known with regard to the mechanism that governs aberrant activation of Ral in bladder cancer, in which Ras mutations are relatively infrequent. Here, we show that Ral was highly activated in invasive bladder cancer cells due to reduced expression of RalGAPa2, the dominant catalytic subunit in bladder, rather than increased expression of RalGEFs. Exogenous expression of wild-type RalGAPa2 in KU7 bladder cancer cells with invasive phenotype, but not mutant RalGAPa2-N1742K lacking RalGAP activity, resulted in attenuated cell migration in vitro and lung metastasis in vivo. Furthermore, genetic ablation of Ralgapa2 promoted tumor invasion in a chemically-induced murine bladder cancer model. Importantly, immunohistochemical analysis of human bladder cancer specimens revealed that lower expression of RalGAPa2 was associated with advanced clinical stage and poor survival of patients. Collectively, these results are highly indicative that attenuated expression of RalGAPa2 leads to disease progression of bladder cancer through enhancement of Ral activity." (Saito et al, Oncogene, 2013)
 "Abstract
Ral small GTPases, consisting of RalA and RalB, are members of the Ras family. Their activity is upregulated by RalGEFs. Since several RalGEFs are downstream effectors of Ras, Ral is activated by the oncogenic mutant Ras. Ral is negatively regulated by RalGAP complexes that consist of a catalytic α1 or α2 subunit and its common partner β subunit and similarly regulate the activity of RalA as well as RalB in vitro. Ral plays an important role in the formation and progression of pancreatic and lung cancers. However, the involvement of Ral in oral squamous cell carcinoma (OSCC) is unclear. In this study, we investigated OSCC by focusing on Ral. OSCC cell lines with high Ral activation exhibited higher motility. We showed that knockdown of RalGAPβ increased the activation level of RalA and promoted the migration and invasion of HSC-2 OSCC cells in vitro. In contrast, overexpression of wild-type RalGAPα2 in TSU OSCC cells attenuated the activation level of RalA and inhibited cell migration and invasion. Real-time quantitative polymerase chain reaction analysis of samples from patients with OSCC showed that RalGAPα2 was downregulated in oral cancer tissues as compared with normal epithelia. Among patients with OSCC, those with a lower expression of RalGAPα2 showed a worse overall survival rate. A comparison of DNA methylation and histone modifications of the RalGAPα2 gene in OSCC cell lines suggested that crosstalk among DNA methylation, histone H4Ac, and H3K27me2 was involved in the downregulation of RalGAPα2. Thus, activation of Ral GTPase by downregulation of RalGAP expression via a potential epigenetic mechanism may enhance OSCC progression.
Keywords:." (Gao et al, J Dentarl Res, 2019)
"Abstract
Ral small GTPases, consisting of RalA and RalB, are members of the Ras family. Their activity is upregulated by RalGEFs. Since several RalGEFs are downstream effectors of Ras, Ral is activated by the oncogenic mutant Ras. Ral is negatively regulated by RalGAP complexes that consist of a catalytic α1 or α2 subunit and its common partner β subunit and similarly regulate the activity of RalA as well as RalB in vitro. Ral plays an important role in the formation and progression of pancreatic and lung cancers. However, the involvement of Ral in oral squamous cell carcinoma (OSCC) is unclear. In this study, we investigated OSCC by focusing on Ral. OSCC cell lines with high Ral activation exhibited higher motility. We showed that knockdown of RalGAPβ increased the activation level of RalA and promoted the migration and invasion of HSC-2 OSCC cells in vitro. In contrast, overexpression of wild-type RalGAPα2 in TSU OSCC cells attenuated the activation level of RalA and inhibited cell migration and invasion. Real-time quantitative polymerase chain reaction analysis of samples from patients with OSCC showed that RalGAPα2 was downregulated in oral cancer tissues as compared with normal epithelia. Among patients with OSCC, those with a lower expression of RalGAPα2 showed a worse overall survival rate. A comparison of DNA methylation and histone modifications of the RalGAPα2 gene in OSCC cell lines suggested that crosstalk among DNA methylation, histone H4Ac, and H3K27me2 was involved in the downregulation of RalGAPα2. Thus, activation of Ral GTPase by downregulation of RalGAP expression via a potential epigenetic mechanism may enhance OSCC progression.
Keywords:." (Gao et al, J Dentarl Res, 2019)